CommunityData:Hyak: Difference between revisions
No edit summary |
|||
| (134 intermediate revisions by 9 users not shown) | |||
| Line 1: | Line 1: | ||
[https://hyak.uw.edu/ Hyak] is the University of Washington's high performance computing (HPC) system. The CDSC has purchased a number of "nodes" on this system, which you will have access to as a member of the group. | |||
To use Hyak, you must first have a UW NetID, access to Hyak, and a two factor authentication token which you will need as part of [[CommunityData:Hyak setup|getting setup]]. The following links will be useful. | |||
* [[CommunityData:Klone]] [[CommunityData:Klone Quick Reference]] (for the new hyak nodes). | |||
* [[CommunityData:Hyak setup]] [[CommunityData:Hyak Quick Reference]] | |||
* [[CommunityData:Hyak software installation]] | |||
* [[CommunityData:Hyak Spark]] | |||
* [[CommunityData:Hyak Mox migration]] | |||
* [[CommunityData:Hyak Ikt (Deprecreated)]] | |||
* [[CommunityData:Hyak Datasets]] | |||
There are a number of other sources of documentation beyond this wiki: | |||
* [http://wiki.hyak.uw.edu Hyak User Documentation] | |||
== General Introduction to Hyak == | |||
The UW Research Computing Club has put together [https://depts.washington.edu/uwrcc/getting-started-2/hyak-training/ this excellent 90 minute training video] that introduces Hyak. It's probably a good place to start for anybody trying to get up-and-running on Hyak. | |||
== Setting up SSH == | == Setting up SSH == | ||
| Line 16: | Line 24: | ||
I've added the following config to the file <code>~/.ssh/config</code> on my laptop (you will want to change the username): | I've added the following config to the file <code>~/.ssh/config</code> on my laptop (you will want to change the username): | ||
Host hyak | Host hyak klone.hyak.uw.edu | ||
User | User '''<YOURNETID>''' | ||
HostName | HostName klone.hyak.uw.edu | ||
ControlPath ~/.ssh/master-%r@%h:%p | ControlPath ~/.ssh/master-%r@%h:%p | ||
ControlMaster auto | ControlMaster auto | ||
| Line 24: | Line 32: | ||
Compression yes | Compression yes | ||
{{Note}} If your SSH connection becomes stale or disconnected (e.g., if you change networks) it may take some time for the connection to time out. Until that happens, any connections you make to hyak will silently hang. If your connections to ssh hyak are silently hanging but your Internet connection seems good, look for ssh processes running on your local machine with: | |||
ps ax|grep hyak | ps ax|grep hyak | ||
If you find any, kill them with <code>kill <PROCESSID></code>. Once that is done, you should have no problem connecting to Hyak. | If you find any, kill them with <code>kill '''<PROCESSID>'''</code>. Once that is done, you should have no problem connecting to Hyak. | ||
== | === X11 forwarding === | ||
{{notice|This is likely only applicable if you are a Linux user}} | |||
You may also want to add these two lines to your Hyak <code>.ssh/config</code> (indented under the line starting with "Host"): | |||
ForwardX11 yes | |||
ForwardX11Trusted yes | |||
These lines will mean that if you have "checked out" an interactive machine, you can ssh from your computer to Hyak and then directly through an addition hop to the machine (like ssh n2347). Those ForwardX11 lines means if you graph things on this session, they will open on your local display. | |||
== Connecting to Hyak == | |||
To connect to Hyak, you now only need to do: | |||
ssh hyak | |||
It will prompt you for your UWNetID's password. Once you type in your password, you will have to respond to a [https://itconnect.uw.edu/security/uw-netids/2fa/ 2-factor authentication request]. | |||
== Setting up your Hyak environment == | |||
Everybody who uses Hyak as part of our group '''must''' add the following line to their <code>~/.bashrc</code> file on Hyak: | |||
<source lang='bash'> | |||
source /gscratch/comdata/env/cdsc_klone_bashrc | |||
</source> | |||
If you don't have a preferred terminal-style text editor, you might start with nano -- <code>nano ~/.bashrc</code>, arrow down, paste in the 'source....' text from above, then ^O to save and ^X to exit. You'll know you were successful when you type <code>more ~/.bashrc</code> and see the 'source....' line at the bottom of the file. Copious information about use of a terminal-style text editor is available online -- common options include nano (basic), emacs (tons of features), and vim (fast). | |||
This line will load scripts that will initialize a good data science environment and set the [[:wikipedia:umask|umask]] so that the files and directories you create are readable by others in the group. '''Please do this immediately before you do any other work on Hyak.''' When you are done, you can reload the shell by logging out and back into Hyak or by running <code lang="bash">exec bash</code>. | |||
== Using the CDSC Hyak Environment == | |||
=== Storing Files === | |||
By default you have access to a home directory with a relatively small quota. There are several dozen terabytes of CDSC-allocated storage in <code>/gscratch/comdata/</code> and you should explore that space. Typically we download | |||
large datasets to <code>/gscratch/comdata/raw_data</code> (see [[#New datasets|the section on new datasets]] below), processed data in <code>/gscratch/comdata/output</code>, and personal workspaces with the need for large data storage in <code>/gscratch/comdata/users/'''<YOURNETID>'''</code>. | |||
=== Basic Commands === | |||
Once you have loaded load modern versions of R and Python and places Spark in your environment. It also provides a number of convenient commands for interacting with the SLURM HPC system for checking out nodes and monitoring jobs. Particularly important commands include | |||
any_machine | |||
which attempts to check out a supercomputing node. | |||
big_machine | |||
Requests a node with 240GB of memory. | |||
build_machine | |||
Checks out a build node which can access the internet and is intended to be used to install software. | |||
ourjobs | |||
Prints all the running jobs by people in the group. | |||
myjobs | |||
Displays jobs by members of the group. | |||
Read the files in <code>/gscratch/comdata/env</code> to see how these commands are created (or run <code>which</code>) as well as other features not documented here. | |||
=== Anaconda === | |||
We recently switched to using Anaconda to manage Python on Hyak. Anaconda comes with the `conda` tool for managing python packages and versions. Multiple python environments can co-exist in a single Anaconda installation, this allows different projects to use different versions of Python or python packages, which can be useful for maintaining projects that use old versions. | |||
By default, our shared setup loads a conda environment called `minimal_ds` that provides recent versions of python packages commonly used in data science workflows. This is probably a good setup for most use-cases, and allows everyone to use the same packages, but it can be even better to create different environments for each project. See the [https://docs.conda.io/projects/conda/en/latest/user-guide/tasks/manage-environments.html#creating-an-environment-with-commands anaconda documentation for how to create an environment]. | |||
To learn how to install Python packages, see the [[CommunityData:Hyak software installation#Python packages|Python packages installation instructions]] on this wiki. | |||
== | === SSH into compute nodes === | ||
The [https://wiki.cac.washington.edu/display/hyakusers/Hyak_ssh hyak wiki] has instructions for how to enable ssh within hyak. Reproduced below: | |||
You should be able to ssh from the login node to a compute node without giving a password. If it does not work then do below steps: | |||
# <code>ssh-keygen</code> then press enter for each question. This will ensure default options. | |||
# <code>cd ~/.ssh</code> | |||
# <code>cat id_rsa.pub >> authorized_keys</code> | |||
== Running Jobs on Hyak == | |||
When you first log in to Hyak, you will be on a "login node". These are nodes that have access to the Internet, and can be used to update code, move files around, etc. They should not be used for computationally intensive tasks. To actually run jobs, there are a few different options, described in detail [http://wiki.cac.washington.edu/display/hyakusers/Mox_scheduler in the Hyak User documentation]. Following are basic instructions for some common use cases. | |||
=== Interactive nodes === | |||
Interactive nodes are systems where you get a <code>bash</code> shell from which you can run your code. This mode of operation is conceptually similar to running your code on your own computer, the difference being that you have access to much more CPU and memory. To check out an interactive node, run the <code>big_machine</code> or <code>any_machine</code> command from your login shell. Before running these commands, you will want to be in a [[CommunityData:Tmux|<code>tmux</code>]] or <code>screen</code> session so that you can start your job, and log off without having to worry about your job getting terminated. | |||
{{note}} At a given point of time, unless you are using the <code>ckpt</code> (formerly the <code>bf</code>) queue, our entire group can collectiveley have one instance of <code>big_machine</code> and three instances of <code>any_machine</code> running at the same time. You may need to coordinate over IRC if you need to use a specific node for any reason. | |||
=== Killing jobs on compute nodes === | |||
The Slurm scheduler provides a command called [https://slurm.schedmd.com/scancel.html scancel] to terminate jobs. For example, you might run <tt>queue_state</tt> from a login node to figure out the ID number for your job (let's say it's 12345), then run <tt>scancel --signal=TERM 12345</tt> to send a SIGTERM signal or <tt>scancel --signal=KILL 12345</tt> to send a SIGKILL signal that will bring job 12345 to an end. | |||
=== Parallelization Tips === | |||
The nodes on Mox have 28 CPU cores. Our nodes on Klone have 40. These may help in speeding up your analysis ''significantly''. If you are using R functions such as <code>lapply</code>, there are parallelized equivalents (e.g. <code>mclappy</code>) which can take advantage of all the cores and give you a 2800% or (4000)% boost! However, something to be aware of here is your code's memory requirement—if you are running 28 processes in parallel, your memory needs can also go up to 28x, which may be more than the ~200GB that the <code>big_machine</code> node on mox will have. In such cases, you may want to dial down the number of CPU cores being used—a way to do that globally in your code is to run the following snippet of code before calling any of the parallelized functions. | |||
If you find yourself doing this often, consider if it is possible to reduce your memory usage via streaming, databases (like sqlite; parquet files; or duckdb), or lower-precision data types (i.e., use 32bit or even 16bit floating point numbers instead of the standard 64bit). | |||
<source lang="r"> | |||
library(parallel) | |||
options(mc.cores=20) ## tell the mc* functions to use 20 cores unless otherwise specified | |||
mcaffinity(1:20) | |||
</source> | |||
More information on parallelizing your R code can be found in the [https://stat.ethz.ch/R-manual/R-devel/library/parallel/doc/parallel.pdf <code>parallel</code> package documentation]. | |||
=== Using the Checkpoint Queue === | |||
Hyak has a special way of scheduling jobs using the '''checkpoint queue'''. When you run jobs on the checkpoint queue, they run on someone else's hyak node that they aren't using right now. This is awesome as it gives us a huge amount of free (as in beer) computing. But using the checkpoint queue does take some effort, mainly because your jobs can get killed at any time if the owner of the node checks it out. So if you want to run a job for more than a few minutes on the checkpoint queue it will need to be able to "checkpoint" by saving it's state periodically and then restarting. | |||
==== Starting a checkpoint queue job ==== | |||
To start a checkpoint queue job we'll use <code>sbatch</code> instead of srun. See the [https://slurm.schedmd.com/sbatch.html documentation] for a refresher starting hpc jobs using sbatch. | |||
To request a job on the checkpoint queue put the following in the top of your <code>sbatch</code> script. | |||
#SBATCH --export=ALL | |||
#SBATCH --account=comdata-ckpt | |||
#SBATCH --partition=ckpt | |||
== New Datasets == | |||
If you want to download a new dataset to Hyak you should first check to ensure that is enough space on the current allocation (e.g., with <code>cat /gscratch/comdata/usage_report.txt</code>. If there is not enough space in our allocation, contact [[Mako]] about getting our allocation increased. It should be fast and easy. | |||
If there is enough space, you should download data to <code>/gscratch/comdata/raw_data/YOURNEWDATASET</code>. | |||
Once you have finished downloading, you should set all the files you have downloaded as read only to prevent people from accidently creating new files, overwriting data, etc. You can do that with the following commands: | |||
= | <syntaxhighlight lang='bash'> | ||
$ cd /gscratch/comdata/raw_data/YOURNEWDATASET | |||
$ find . -not -type d -print0 |xargs -0 chmod 440 | |||
$ find . -type d -print0 |xargs -0 chmod 2550 | |||
</syntaxhighlight> | |||
= Tips and Faqs = | |||
== | == 5 productivity tips == | ||
# Find a workflow that works for you. There isn't a standardized workflow for quantitative / computational social science or social computing. People normally develop idiosyncratic workflows around the distinctive tools they know or have been exposed and that meet their diverse needs and tastes. Be aware of how you're spending your time and effort and adopt tools in your workflow that make things easier or more efficient. For example, if you're spending a lot of time typing into the hyak command line, bash-completion and bash-history can help, and a pipeline (see below) might help even more. | |||
# If you find yourself spending time manually rerunning code in a multistage project, learn [https://en.wikipedia.org/wiki/Make_(software) Make] or another pipeline tool. Such tools take some effort but really help you organize, test, and refine your project. Make is a good choice because it is old and incredibly polished and featureful. You don't need to learn every feature, just the basics. Its interface has a different flavor than more recently designed tools which can be a downside. Other positives are that it is language agnostic and can run shell commands. | |||
# [https://slurm.schedmd.com/documentation.html Slurm] the system that you use to access hyak nodes, is also a very powerful system. The hyak team used to maintain a tool called parallel-sql which helped with running a large number of short-running programs. This tool is no longer supported, but [https://slurm.schedmd.com/job_array.html job arrays] are slurm feature that is even better. | |||
# Use the free resources. Job arrays (mentioned above) are great in combination with the [https://wiki.cac.washington.edu/display/hyakusers/Mox_checkpoint checkpoint queue]. The checkpoint (or ckpt) queue runs your jobs on other people's idle nodes. You can access thousands of cores and terabytes of RAM on the checkpoint queue. There are limitations. If the owner of a node wants to use it, they will cancel your job. If this happens, the scheduler will automatically restart it, and it has a maximum total running time (restarts don't reset the clock). Therefore, it is best suited for jobs that can be paused (saved) and restarted. If you can design a script to catch the checkpoint signal, save progress, and restart you will be able to make excellent use of the checkpoint queue. Note that checkpoint jobs get run according to a priority system and if members of our group overuse this resource then our jobs will have lower priority. <br /> There is also virtually [https://hyak.uw.edu/docs/storage/gscratch/ unlimited free storage] on hyak under <code>/gscratch/scrubbed/comdata</code> with the catch that the storage is much slower and that files will be automatically deleted after a short time (currently 21 days). | |||
# Get connected to the hyak team and other hyak users. Hyak isn't perfect and has many recent issues related to the new Klone system. If you run into trouble and it feels like the system isn't working you should email help@uw.edu with a subject line that starts with "hyak:". They are nice and helpful. Other good resources are the [https://mailman12.u.washington.edu/mailman/listinfo/hyak-users mailing list] and if you are a UW student, the [https://depts.washington.edu/uwrcc/getting-started-2/getting-started/ research computing club]. The club has its own nodes, including GPU nodes that only students who join the club can use. | |||
=== | == Common Troubles and How to Solve Them == | ||
=== Help! I'm over CPU quota and Hyak is angry! === | |||
'''Don't panic.''' Everyone has done this at least once. Mako has done it dozens of times. It is a little bit difficult to deal with but can be solved. You are not in trouble. | |||
The usual reason for this to happen is because you've accidentally run something on a login node that ought to be run on a compute node. The solution is to find the badly behaved process and then use kill to kill the process. | |||
If it's a script or command on your commandline, '''Ctrl-c''' to kill it. If you backgrounded it, type <code>fg</code> to foreground it and then '''Ctrl-c'''. But if you ran parallel, you'll need to kill parallel itself. | |||
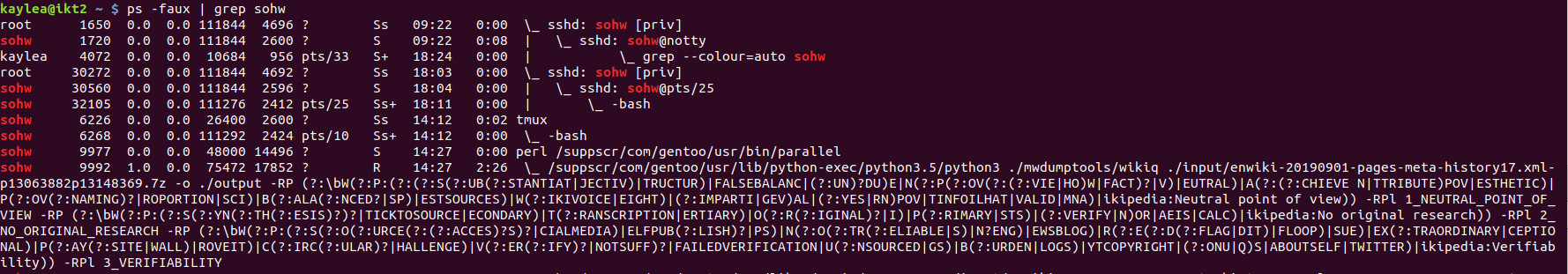
<code>ps -faux | grep <your username></code> will show you all the things you are running (or have someone else run it for you if the spam is so terrible you can't get a command to run). The first column has the usernames, the second column has the process IDs, the last column has the things you're running. | |||
[[File:faux.jpg]] | |||
In the screenshot, the red is the user name being grepped for. At the end of the line the last three entries are the time (in hyak time, type date if you want to compare hyak time to your time), then how much CPU time something has consumed, then a little diagram of parent and child processes. You want parallel (in the example, 9977). | |||
Killing the child process (in the example, 9992) won't likely help because parallel will just go on to the next task you queued up for it. You will need to run something like: <code>kill <process id></code> | |||
=== My R Job is getting Killed === | |||
First, make sure you're running on a compute node (like n2344) or else the int_machine and don't use a --time-min flag -- there seems to be a bug with --time-min where it evicts jobs incorrectly. | |||
Second, see if you can narrow down where in your R code the problem is happening. Kaylea has seen it primarily when reading or writing files, and this tip is from that experience. Breaking the read or write into smaller chunks (if that makes sense for your project) might be all it takes. | |||
Latest revision as of 17:35, 17 October 2024
Hyak is the University of Washington's high performance computing (HPC) system. The CDSC has purchased a number of "nodes" on this system, which you will have access to as a member of the group.
To use Hyak, you must first have a UW NetID, access to Hyak, and a two factor authentication token which you will need as part of getting setup. The following links will be useful.
- CommunityData:Klone CommunityData:Klone Quick Reference (for the new hyak nodes).
- CommunityData:Hyak setup CommunityData:Hyak Quick Reference
- CommunityData:Hyak software installation
- CommunityData:Hyak Spark
- CommunityData:Hyak Mox migration
- CommunityData:Hyak Ikt (Deprecreated)
- CommunityData:Hyak Datasets
There are a number of other sources of documentation beyond this wiki:
General Introduction to Hyak[edit]
The UW Research Computing Club has put together this excellent 90 minute training video that introduces Hyak. It's probably a good place to start for anybody trying to get up-and-running on Hyak.
Setting up SSH[edit]
When you connect to SSH, it will ask you for a key from your token. Typing this in every time you start a connection be a pain. One approach is to create an .ssh config file that will create a "tunnel" the first time you connect and send all subsequent connections to Hyak over that tunnel. Some details in the Hyak documentation.
I've added the following config to the file ~/.ssh/config on my laptop (you will want to change the username):
Host hyak klone.hyak.uw.edu
User <YOURNETID>
HostName klone.hyak.uw.edu
ControlPath ~/.ssh/master-%r@%h:%p
ControlMaster auto
ControlPersist yes
Compression yes
![]() Note: If your SSH connection becomes stale or disconnected (e.g., if you change networks) it may take some time for the connection to time out. Until that happens, any connections you make to hyak will silently hang. If your connections to ssh hyak are silently hanging but your Internet connection seems good, look for ssh processes running on your local machine with:
Note: If your SSH connection becomes stale or disconnected (e.g., if you change networks) it may take some time for the connection to time out. Until that happens, any connections you make to hyak will silently hang. If your connections to ssh hyak are silently hanging but your Internet connection seems good, look for ssh processes running on your local machine with:
ps ax|grep hyak
If you find any, kill them with kill <PROCESSID>. Once that is done, you should have no problem connecting to Hyak.
X11 forwarding[edit]
| This is likely only applicable if you are a Linux user |
You may also want to add these two lines to your Hyak .ssh/config (indented under the line starting with "Host"):
ForwardX11 yes ForwardX11Trusted yes
These lines will mean that if you have "checked out" an interactive machine, you can ssh from your computer to Hyak and then directly through an addition hop to the machine (like ssh n2347). Those ForwardX11 lines means if you graph things on this session, they will open on your local display.
Connecting to Hyak[edit]
To connect to Hyak, you now only need to do:
ssh hyak
It will prompt you for your UWNetID's password. Once you type in your password, you will have to respond to a 2-factor authentication request.
Setting up your Hyak environment[edit]
Everybody who uses Hyak as part of our group must add the following line to their ~/.bashrc file on Hyak:
source /gscratch/comdata/env/cdsc_klone_bashrc
If you don't have a preferred terminal-style text editor, you might start with nano -- nano ~/.bashrc, arrow down, paste in the 'source....' text from above, then ^O to save and ^X to exit. You'll know you were successful when you type more ~/.bashrc and see the 'source....' line at the bottom of the file. Copious information about use of a terminal-style text editor is available online -- common options include nano (basic), emacs (tons of features), and vim (fast).
This line will load scripts that will initialize a good data science environment and set the umask so that the files and directories you create are readable by others in the group. Please do this immediately before you do any other work on Hyak. When you are done, you can reload the shell by logging out and back into Hyak or by running exec bash.
Using the CDSC Hyak Environment[edit]
Storing Files[edit]
By default you have access to a home directory with a relatively small quota. There are several dozen terabytes of CDSC-allocated storage in /gscratch/comdata/ and you should explore that space. Typically we download
large datasets to /gscratch/comdata/raw_data (see the section on new datasets below), processed data in /gscratch/comdata/output, and personal workspaces with the need for large data storage in /gscratch/comdata/users/<YOURNETID>.
Basic Commands[edit]
Once you have loaded load modern versions of R and Python and places Spark in your environment. It also provides a number of convenient commands for interacting with the SLURM HPC system for checking out nodes and monitoring jobs. Particularly important commands include
any_machine
which attempts to check out a supercomputing node.
big_machine
Requests a node with 240GB of memory.
build_machine
Checks out a build node which can access the internet and is intended to be used to install software.
ourjobs
Prints all the running jobs by people in the group.
myjobs
Displays jobs by members of the group.
Read the files in /gscratch/comdata/env to see how these commands are created (or run which) as well as other features not documented here.
Anaconda[edit]
We recently switched to using Anaconda to manage Python on Hyak. Anaconda comes with the `conda` tool for managing python packages and versions. Multiple python environments can co-exist in a single Anaconda installation, this allows different projects to use different versions of Python or python packages, which can be useful for maintaining projects that use old versions.
By default, our shared setup loads a conda environment called `minimal_ds` that provides recent versions of python packages commonly used in data science workflows. This is probably a good setup for most use-cases, and allows everyone to use the same packages, but it can be even better to create different environments for each project. See the anaconda documentation for how to create an environment.
To learn how to install Python packages, see the Python packages installation instructions on this wiki.
SSH into compute nodes[edit]
The hyak wiki has instructions for how to enable ssh within hyak. Reproduced below:
You should be able to ssh from the login node to a compute node without giving a password. If it does not work then do below steps:
ssh-keygenthen press enter for each question. This will ensure default options.cd ~/.sshcat id_rsa.pub >> authorized_keys
Running Jobs on Hyak[edit]
When you first log in to Hyak, you will be on a "login node". These are nodes that have access to the Internet, and can be used to update code, move files around, etc. They should not be used for computationally intensive tasks. To actually run jobs, there are a few different options, described in detail in the Hyak User documentation. Following are basic instructions for some common use cases.
Interactive nodes[edit]
Interactive nodes are systems where you get a bash shell from which you can run your code. This mode of operation is conceptually similar to running your code on your own computer, the difference being that you have access to much more CPU and memory. To check out an interactive node, run the big_machine or any_machine command from your login shell. Before running these commands, you will want to be in a tmux or screen session so that you can start your job, and log off without having to worry about your job getting terminated.
![]() Note: At a given point of time, unless you are using the
Note: At a given point of time, unless you are using the ckpt (formerly the bf) queue, our entire group can collectiveley have one instance of big_machine and three instances of any_machine running at the same time. You may need to coordinate over IRC if you need to use a specific node for any reason.
Killing jobs on compute nodes[edit]
The Slurm scheduler provides a command called scancel to terminate jobs. For example, you might run queue_state from a login node to figure out the ID number for your job (let's say it's 12345), then run scancel --signal=TERM 12345 to send a SIGTERM signal or scancel --signal=KILL 12345 to send a SIGKILL signal that will bring job 12345 to an end.
Parallelization Tips[edit]
The nodes on Mox have 28 CPU cores. Our nodes on Klone have 40. These may help in speeding up your analysis significantly. If you are using R functions such as lapply, there are parallelized equivalents (e.g. mclappy) which can take advantage of all the cores and give you a 2800% or (4000)% boost! However, something to be aware of here is your code's memory requirement—if you are running 28 processes in parallel, your memory needs can also go up to 28x, which may be more than the ~200GB that the big_machine node on mox will have. In such cases, you may want to dial down the number of CPU cores being used—a way to do that globally in your code is to run the following snippet of code before calling any of the parallelized functions.
If you find yourself doing this often, consider if it is possible to reduce your memory usage via streaming, databases (like sqlite; parquet files; or duckdb), or lower-precision data types (i.e., use 32bit or even 16bit floating point numbers instead of the standard 64bit).
library(parallel)
options(mc.cores=20) ## tell the mc* functions to use 20 cores unless otherwise specified
mcaffinity(1:20)
More information on parallelizing your R code can be found in the parallel package documentation.
Using the Checkpoint Queue[edit]
Hyak has a special way of scheduling jobs using the checkpoint queue. When you run jobs on the checkpoint queue, they run on someone else's hyak node that they aren't using right now. This is awesome as it gives us a huge amount of free (as in beer) computing. But using the checkpoint queue does take some effort, mainly because your jobs can get killed at any time if the owner of the node checks it out. So if you want to run a job for more than a few minutes on the checkpoint queue it will need to be able to "checkpoint" by saving it's state periodically and then restarting.
Starting a checkpoint queue job[edit]
To start a checkpoint queue job we'll use sbatch instead of srun. See the documentation for a refresher starting hpc jobs using sbatch.
To request a job on the checkpoint queue put the following in the top of your sbatch script.
#SBATCH --export=ALL #SBATCH --account=comdata-ckpt #SBATCH --partition=ckpt
New Datasets[edit]
If you want to download a new dataset to Hyak you should first check to ensure that is enough space on the current allocation (e.g., with cat /gscratch/comdata/usage_report.txt. If there is not enough space in our allocation, contact Mako about getting our allocation increased. It should be fast and easy.
If there is enough space, you should download data to /gscratch/comdata/raw_data/YOURNEWDATASET.
Once you have finished downloading, you should set all the files you have downloaded as read only to prevent people from accidently creating new files, overwriting data, etc. You can do that with the following commands:
$ cd /gscratch/comdata/raw_data/YOURNEWDATASET
$ find . -not -type d -print0 |xargs -0 chmod 440
$ find . -type d -print0 |xargs -0 chmod 2550
Tips and Faqs[edit]
5 productivity tips[edit]
- Find a workflow that works for you. There isn't a standardized workflow for quantitative / computational social science or social computing. People normally develop idiosyncratic workflows around the distinctive tools they know or have been exposed and that meet their diverse needs and tastes. Be aware of how you're spending your time and effort and adopt tools in your workflow that make things easier or more efficient. For example, if you're spending a lot of time typing into the hyak command line, bash-completion and bash-history can help, and a pipeline (see below) might help even more.
- If you find yourself spending time manually rerunning code in a multistage project, learn Make or another pipeline tool. Such tools take some effort but really help you organize, test, and refine your project. Make is a good choice because it is old and incredibly polished and featureful. You don't need to learn every feature, just the basics. Its interface has a different flavor than more recently designed tools which can be a downside. Other positives are that it is language agnostic and can run shell commands.
- Slurm the system that you use to access hyak nodes, is also a very powerful system. The hyak team used to maintain a tool called parallel-sql which helped with running a large number of short-running programs. This tool is no longer supported, but job arrays are slurm feature that is even better.
- Use the free resources. Job arrays (mentioned above) are great in combination with the checkpoint queue. The checkpoint (or ckpt) queue runs your jobs on other people's idle nodes. You can access thousands of cores and terabytes of RAM on the checkpoint queue. There are limitations. If the owner of a node wants to use it, they will cancel your job. If this happens, the scheduler will automatically restart it, and it has a maximum total running time (restarts don't reset the clock). Therefore, it is best suited for jobs that can be paused (saved) and restarted. If you can design a script to catch the checkpoint signal, save progress, and restart you will be able to make excellent use of the checkpoint queue. Note that checkpoint jobs get run according to a priority system and if members of our group overuse this resource then our jobs will have lower priority.
There is also virtually unlimited free storage on hyak under/gscratch/scrubbed/comdatawith the catch that the storage is much slower and that files will be automatically deleted after a short time (currently 21 days). - Get connected to the hyak team and other hyak users. Hyak isn't perfect and has many recent issues related to the new Klone system. If you run into trouble and it feels like the system isn't working you should email help@uw.edu with a subject line that starts with "hyak:". They are nice and helpful. Other good resources are the mailing list and if you are a UW student, the research computing club. The club has its own nodes, including GPU nodes that only students who join the club can use.
Common Troubles and How to Solve Them[edit]
Help! I'm over CPU quota and Hyak is angry![edit]
Don't panic. Everyone has done this at least once. Mako has done it dozens of times. It is a little bit difficult to deal with but can be solved. You are not in trouble.
The usual reason for this to happen is because you've accidentally run something on a login node that ought to be run on a compute node. The solution is to find the badly behaved process and then use kill to kill the process.
If it's a script or command on your commandline, Ctrl-c to kill it. If you backgrounded it, type fg to foreground it and then Ctrl-c. But if you ran parallel, you'll need to kill parallel itself.
ps -faux | grep <your username> will show you all the things you are running (or have someone else run it for you if the spam is so terrible you can't get a command to run). The first column has the usernames, the second column has the process IDs, the last column has the things you're running.
In the screenshot, the red is the user name being grepped for. At the end of the line the last three entries are the time (in hyak time, type date if you want to compare hyak time to your time), then how much CPU time something has consumed, then a little diagram of parent and child processes. You want parallel (in the example, 9977).
Killing the child process (in the example, 9992) won't likely help because parallel will just go on to the next task you queued up for it. You will need to run something like: kill <process id>
My R Job is getting Killed[edit]
First, make sure you're running on a compute node (like n2344) or else the int_machine and don't use a --time-min flag -- there seems to be a bug with --time-min where it evicts jobs incorrectly.
Second, see if you can narrow down where in your R code the problem is happening. Kaylea has seen it primarily when reading or writing files, and this tip is from that experience. Breaking the read or write into smaller chunks (if that makes sense for your project) might be all it takes.