CommunityData:Build papers
When creating LaTeX documents, the final PDF output must be built from an input file. For many of our projects, this process is even more complicated and we use Makefiles to manage more complex workflows. This document is intended to give an overview of the basic process and to identify good practices for quantitative projects.
This sketch illustrates an overall setup: 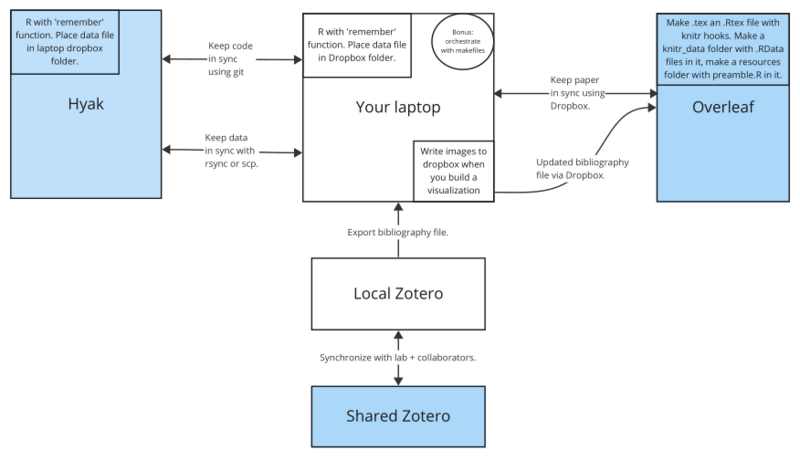
(a PDF version if you prefer) to see how these pieces can fit together.
Project Creation[edit]
Follow the directions for installing the LaTeX templates. Once they are installed, you can run something like:
cd ~/Projects new_knitr_document project_name
This will create a new project called project_name.
This project will include a Makefile and all of the pieces needed to create a document.
Knitr[edit]
Zotero[edit]
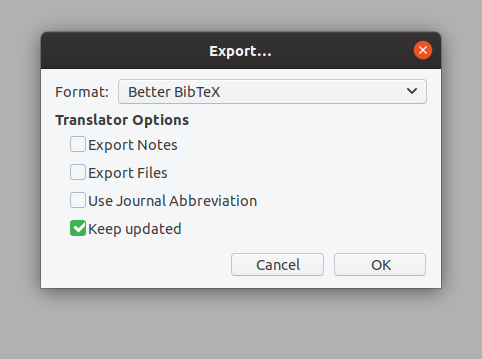
The next step is to set up your references. Often, this means creating a new directory in Zotero. Follow the directions on the CommunityData:Zotero page. You should export your Zotero directory as Better BibLaTeX to the refs.bib file, and check "Keep updated" for Zotero to automatically update that file whenever the directory changes.
Making the paper[edit]
- The first time you're building the paper, you can just run
makeormake all. After that, you probably want to runmake clean; make all. This should work whether you're using an .Rtex (knitr) or .tex (LaTeX) file.
If you need to output a Word document (e.g., for a journal submission), see the LaTeX to Word page.
Git[edit]
At this point, you should put your project into git. See CommunityData:Git for instructions.
Overleaf[edit]
If you're using Overleaf for your papers, that's great. Many of us use Overleaf at some point in our writing process. This section describes a reasonable workflow for putting papers on Overleaf.
Dropbox Sync[edit]
Overleaf is actually really good at allowing for a combination online/offline workflow, and keeping documents in sync. Most of us have this set up through Dropbox sync. In your Overleaf account settings you can set this up. Once you do that, all of your Overleaf projects will appear in ~/Dropbox/Apps/Overleaf/.
If you don't want to fiddle with account settings on Overleaf, you can also simply:
- Get Better Bibtex on your Desktop Zotero app.
- Export your collection/library to "auto" update from Zotero (See this below) to an auto-syncing Dropbox folder. You will probably need to have the DropBox Desktop app.
- To avoid exporting fields in Zotero that you don't want, like ISBN of a conference paper, go to Edit > Preference > Export > Fields, and type the fields you don't want in the appropriate box. Do this *before* you export your auto-updating collection/library.
- Follow the instructions here to link the .bib file that is auto-exported by Better Bibtex and synced to Dropbox to your Overleaf, but note the changes for Google Drive: https://www.overleaf.com/learn/how-to/How_can_I_upload_files_from_Google_Drive%3F
- For DropBox-synced files:
- On Overleaf, upload a new file by selecting "From external URL". Copy and paste the URL from the link to the file that DropBox gives you for the file you saved your collection/library as. It will look like
https://www.dropbox.com/s/[NUMBERS_AND_LETTERS]/[FILENAME].bib?dl=0. - Replace
dl=0withraw=1 - Enter the "File Name In This Project" for the Overleaf project (e.g. refs.bib). This is what you'll refer to your bib file as in the TeX, and hit Create.
- On Overleaf, upload a new file by selecting "From external URL". Copy and paste the URL from the link to the file that DropBox gives you for the file you saved your collection/library as. It will look like
- When there are changes made on Zotero, note that you need to "refresh" the file on Overleaf to get the updated/synced file.
Creating a Project[edit]
Overleaf supports the entire pipeline that we typically use when building TeX papers from a local computer, including knitr and Makefiles. Therefore, the best way to put a complicated project onto Overleaf is to create it on your computer first. You can do this by going to the Overleaf directory in Dropbox and using the new_knitr_document command. This will both give you a new document with all of the needed resources and will create a new project on Overleaf
Data and Code[edit]
You should not put your entire project on Overleaf. If you follow the suggested directory structure below, then Overleaf should just contain the paper/ directory. The code and data should live somewhere else, typically.
The one problem with this is that you likely want the whole project, including the paper, in the same place, and updated in a git repository. One approach to this is to include an option in the Makefile which will copy the Dropbox version into the canonical directory.
The bibliography file[edit]
The Best Practice is still to use Zotero to manage your bibliography. If you have installed Better Bibtex (and if you haven't, then you should!), then you can export your bibliography and keep it updated. You right-click on the collection you want to export, choose the format (likely Better BibTex), and select "keep updated"
You should export it into the refs.bib file in your Dropbox Overleaf project, and then Better Bibtex and Dropbox will keep it updated on Overleaf for you.
Style notes and more details (add as needed)[edit]
- The typical directory structure that I use is something like:
/code
01_script.py
02_script.py
03_analysis.R
README
/data
/raw_data
raw_data_file.csv
measures.csv
analyzed_data.RData
/paper
/data
analyzed_data.RData (symlinked)
/figures (any figures not created with knitr)
paper.Rtex
The code all lives in one place, with a README that explains what the pieces do. Don't underestimate the importance of a README and well-commented code. Your future self will thank you.
The data lives in another place, with raw_data which is read-only (and may or may not be stored in git, depending on the size). This raw data is used to create measures (and possibly some intermediate files). Analysis is done on the measures file, and stored in an RData file.
It is tempting to put as much of the analysis workflow as possible into the .Rtex file but these files can quickly become difficult to compile and unwieldy. It's often better to do the heavy lifting in a separate script, import an .RData file in the paper, and do things like creating plots and running simple statistical tests in the paper itself.
- You may need to use natbib for some journal submission styles. The stackexchange page linked above helps to explain how to do that (but if anyone wants to give more detailed instructions, please do!).